Co-Project leader: Dr. Matthias Lange (IPK)
Email:Staff:
- Dr. Daniel Arend
- Elena Rey-Mazón
The aim of WP II.12 (IT-solutions for public data storage and access), which is schematically presented in Figure 1, is developing a central data infrastructure for the long-term storage of quality-assured, integrated, multi-omics project data secured according to FAIR criteria. The necessary data structures, as well as the interfaces and integration strategies, will be designed and implemented according to the project requirements. This includes customized data import, management and programmatic interfaces. A data curator monitors data quality according to the specifications of identifiers, vocabularies, formats and scale
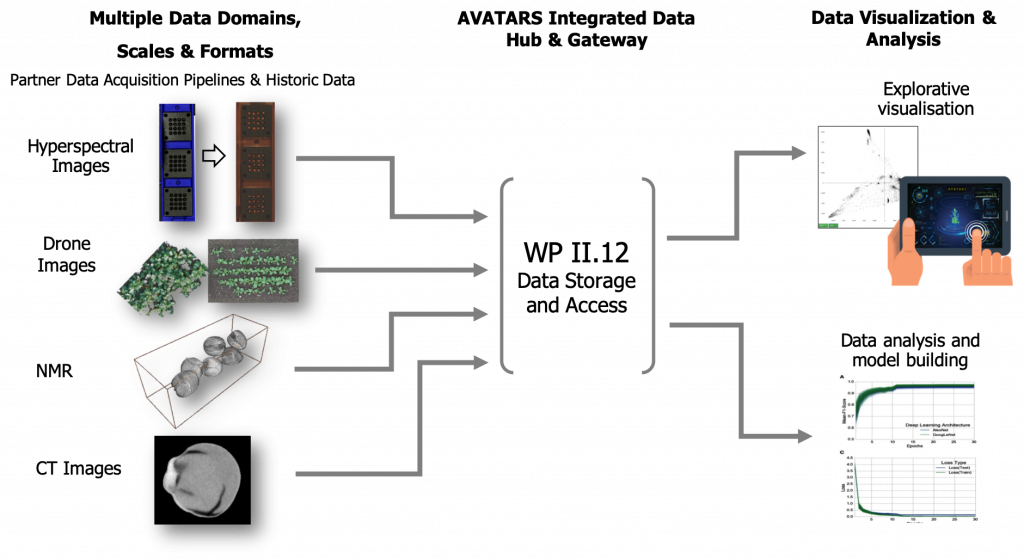
This data is provided in an interoperable and integrated data warehouse infrastructure. The general concept of this infrastructure is shown in Figure 2. A central relational database system takes care of managing granular, operational data from experiments. Binary data is stored in an open and harmonized format in a file system. The project partner IPK will host both. WP II.12 will provide appropriate data storage capacity using a multi-tier file storage management system.
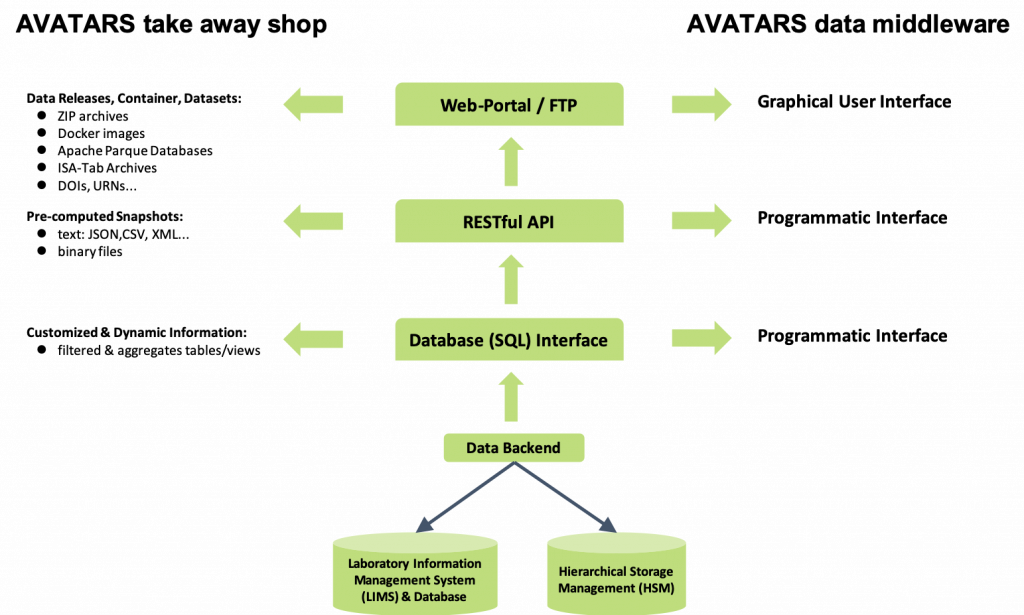
The result will be scalable and homogeneous access to all project data via a provided web portal. This will be achieved either by packaging and delivering the data for edge computing or by providing online access to the IPK infrastructure for client-server infrastructures.
Publications
The research data life cycle is crucial, spanning from project planning to data publishing. In the past, researchers handled all phases, often with limited assistance. However, advancements in sequencing and automation have unleashed a data deluge, surpassing researchers' expertise and infrastructure. This raises the risk of data loss. IPK Gatersleben, with vast germplasm collections and expertise in crop plant research data management, demonstrates how data stewards can tackle challenges in modern research. Through concrete examples and best practices from plant phenotyping, we explore the expertise and skills required for a transformative digital journey in progressive research.
Arend, D., Psaroudakis, D., Memon, J. A., Rey‐Mazón, E., Schüler, D., Szymanski, J. J., Scholz, U., Junker, A., & Lange, M. (2022). From data to knowledge – big data needs stewardship, a plant phenomics perspective. In The Plant Journal (Vol. 111, Issue 2, pp. 335–347). Wiley. https://doi.org/10.1111/tpj.15804
Integrative bioinformatics combines data from different domains to enable cross-domain analysis, unlocking insights not achievable with single-domain approaches. For instance, it allows us to explore the relationships between genotypes, phenotypes, and environmental contexts. Effective data management and machine-readable access are vital to facilitate this process. This chapter focuses on the data life cycle, covering steps from planning to sharing and reusing data. The aim is to make research results accessible to the public, adhering to the FAIR principles of Findability, Accessibility, Interoperability, and Reusability.
Arend, D., Beier, S., König, P., Lange, M., Memon, J. A., Oppermann, M., Scholz, U., & Weise, S. (2022). From Genotypes to Phenotypes: A Plant Perspective on Current Developments in Data Management and Data Publication. In Integrative Bioinformatics (pp. 11–43). Springer Singapore. https://doi.org/10.1007/978-981-16-6795-4_2