Co-Project leader: Dr. Evgeny Gladilin
Email:Multimodal images of seeds (such as 2D cross-section and 3D MRI data) have to be aligned, segmented and normalized for subsequent analysis and consistent visualization. Thereby, different seeds can, in general, exhibit relative rigid as well as non-rigid transformations that have to be calculated in an automated manner. To address this task, reliable methods for rigid and non-rigid alignment of 2D/3D images are investigated. Based on the presently available exemplary images of Brassica napus seeds, first feasibility studies towards alignment and normalization of different seed images were performed. Alignment of multiple seed images provides the basis for consistent statistical analysis of spectral characteristics and related molecular properties of different seed organs such as cotyledons and radicule. Normalized images of seeds will be subsequently registered in the AVATARS data hub at IPK and provided to cooperating groups that deal with combined image/omics data analysis and visualization.
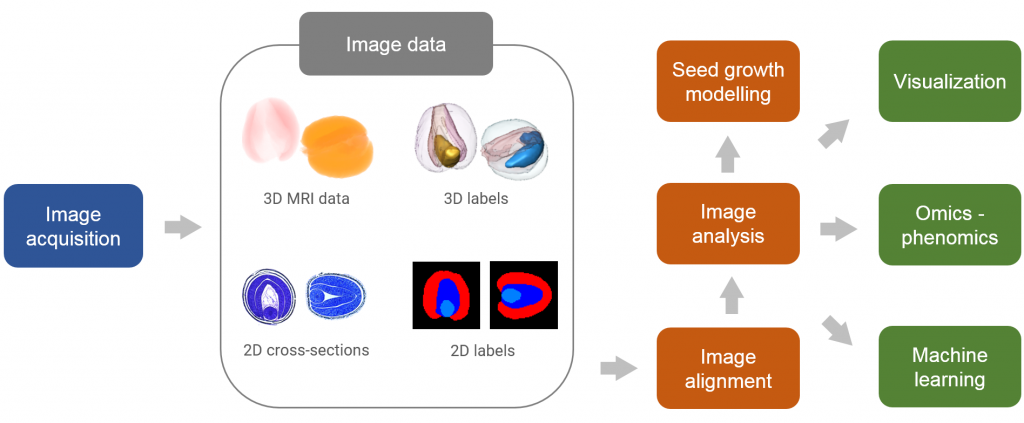
In addition to the alignment of different seed images from the same developmental stage, tracking changes in time-series of seed images provides insights into the dynamics of seed organ growth in different seed accessions. For this purpose, time-series of segmented and normalized 3D MRI images of multiple seeds will be used to generate statistical models of 3D seed morphology and to simulate dynamics of tissue/organ formation during seed development. Key parameters of seed development, such as tissue growth rates, principle stretches, etc., derived from computer simulation, will be used to extend conventional phenotypic descriptors (e.g., seed volume, shape) in genotype/phenotype studies.
Further activities related to the above research tasks include cooperation with research groups dealing with microscopic imaging, data mining, visualization as well as the specification of data access patterns for the further development of performant interfaces to the AVATARS data hub for consistent data storage and sharing within the project consortium.